4. Logging the Simulation¶
4.1. Logging system¶
TREvoSim has a versatile logging system which allows the user to define outputs in a range of formats required for phylogenetic inference packages, provided these allow plain text inputs. Clicking on the output button of the toolbar will launch the output dialogue:
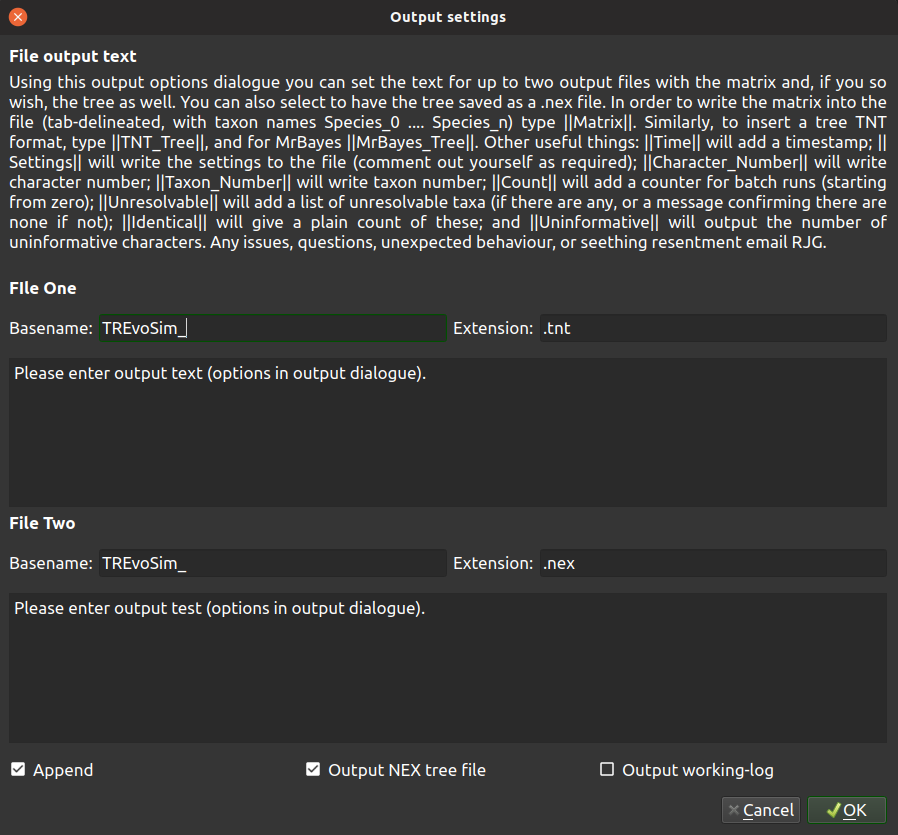
This provides the options for two custom log files, which are placed, after a run, within the TREvoSim_output folder created on the save path. Basename defines the start of the filename, which then either includes a number, which iterates with subsequent runs when output is not set to append, or _batch if the outputs are set to append and a batch is being run. The file extension is also defined here. The two text boxes then allow custom file content to be written: text is written as provided to the file - for example with run instructions or program commands - and any of the keywords below (note this is case sensitive) included within two vertical bar ( | ) symbols are replaced as a file is written.
4.2. Keywords¶
Keywords within two vertical bars ( e.g. |Matrix| ) are replaced as a file is written as follows:
| Matrix: | This is replaced with the matrix from the run. |
|---|---|
| TNT_Tree: | This writes a tree, if required, in TNT format (i.e. only brackets and terminal labels), e.g. |
(((((00 (((((((((((((((05 20) 19) 18) 17) 16) 15) (14 ((((21 ((25 ((((27 31) 30) 29) 28)) 26)) 24) 23) 22))) 13) 12) 11) 10) 09) 08) 07) 06)) 04) 03) 02) 01)
| MrBayes_Tree: | This writes a tree in standard Newick format, including branch lengths (these are based on iteration number throughout), e.g. |
|---|
(S_01:13,(S_02:8,(S_03:13,(S_04:32,((S_06:4,(S_07:10,(S_08:6,(S_09:5,(S_10:10,(S_11:6,(S_12:6,(S_13:2,(((S_22:11,(S_23:12,(S_24:9,((S_26:28,((S_28:37,(S_29:2,(S_30:2,(S_31:1,S_27:1):1):38):23):20,S_25:12):31):41,S_21:31):24):27):1):70,S_14:13):42,(S_15:21,(S_16:2,(S_17:11,(S_18:2,(S_19:10,(S_20:14,S_05:25):7):1):4):2):1):2):1):13):58):6):2):2):5):2):36,S_00:20):3):1):3):7):85
Note The mechanism used for tree writing differs between the above requests - the tree topology is the same, but the taxon order differs.
| Time: | Adds a timestamp. |
|---|---|
| Settings: | Writes the settings to the file (this is provided as a useful way to record, with any output data, the set up for TREvoSim for any given run). |
| Character_Number: | |
| This outputs the character number. | |
| Taxon_Number: | Writes taxon number. |
| Count: | This is replaced with a counter for batch runs; incrementing by one using C++ numbering (i.e. starting from zero). |
| Unresolvable: | This prints a list of unresolvable taxa (or a notice that there are none if required). |
| Uninformative: | Writes the number of uninformative characters. |
So, for example, the following would output a block of text that could be run as a macro in tnt:
NSTATES nogaps;
xread
'Written on ||Time|| Variables: ||Settings||'
||Character_Number|| ||Taxon_Number||
||Matrix||
;
piwe-;
keep 0; hold 100000;
rseed *;
xmult = level 10; bbreak;
export - ||Count||_POUT.nex;
xwipe;
There are three further options at the base of the dialogue:
| Append: | When checked, in batch mode, all the outputs are appended onto a single file, allowing them to be run as a single analysis in - for example - TNT and MrBayes. |
|---|---|
| Output NEX tree file: | |
| This outputs a tree for each run in a standard nexus format with the tree and translate block, as well as a comment with the settings of the run written to it. These do not append, but each file has the run number at the end of the file name. | |
| Output working log: | |
| When this is checked, TREvoSim outputs a text file outlining many of the steps each iteration, such as the state of the playing field, the environment, and the processes the software is going through. This helps understand and fact check any given run, but for significant playing field sizes, taxon numbers, or character numbers, it creates a significant (10s - 100s of MB) text file. | |
Should any other output options be required, please file a feature request. Keywords are not case sensitive.